
engineering & technology publications
ISSN 1759-3433
PROCEEDINGS OF THE FIFTH INTERNATIONAL CONFERENCE ON ENGINEERING COMPUTATIONAL TECHNOLOGY
Probabilistic Regulatory Networks: Modelling Genetic Networks
1Department of Mathematic-Physics, Cayey,
2Department of Mathematics and Computer Sciences, Rio Piedras
University of Puerto Rico, United States of America
If ![]() is a set of selection probabilities we denote by
is a set of selection probabilities we denote by ![]() the
characteristic function over
the
characteristic function over ![]() ,
,
![]() such that
such that ![]() , if
, if ![]() and
and ![]() . Let
. Let
![]() and
and
![]() be two PRN. A map
be two PRN. A map
![]() is an
is an ![]() -homomorphism
from
-homomorphism
from
![]() to
to
![]() , if for a fixed real number
, if for a fixed real number
![]() , and for all
, and for all ![]() there exists a
there exists a ![]() , such that
for all u, v in
, such that
for all u, v in
![]() ,
,
![]()
![]() and
and
![]() If
If
![]() is a bijective map, and
is a bijective map, and
![]() , for all
, for all ![]() ,
, ![]() , u,
and v in
, u,
and v in
![]() ; then
; then ![]() is an isomorphism. If
is an isomorphism. If
![]() and
and ![]() are
bijective
are
bijective ![]() -homomorphisms, then the transition matrices
-homomorphisms, then the transition matrices
![]() and
and ![]() satisfy the condition
max
satisfy the condition
max
![]() ,
for all
,
for all ![]() ; u, v, in
; u, v, in
![]() , where
, where
![]() , and
, and
![]() .
This property determines a correspondence between the Markov chains of
these two networks. Here, we introduce the concept of two similar
time discrete Markov chains (TDMC).
Two TDMC of the same size
.
This property determines a correspondence between the Markov chains of
these two networks. Here, we introduce the concept of two similar
time discrete Markov chains (TDMC).
Two TDMC of the same size
![]() :
:
![]() , and
, and
![]() , are
, are ![]() -similar or
-similar or ![]() -isomorphic if a
-isomorphic if a
![]() exists that is small enough, such that
exists that is small enough, such that
![]() satisfies that
satisfies that
![]() for all m, where
for all m, where
![]() is the characteristic function.
is the characteristic function.
For example: suppose we have
two genes with two values that we denote as usual ![]() , that is
this PRN is a very simple PBN. The set of boolean functions F is
the following:
, that is
this PRN is a very simple PBN. The set of boolean functions F is
the following:
![]()
![]() and the probabilities are
and the probabilities are
![]() . In Figure 1, the PBN
. In Figure 1, the PBN
![]() ,
its state space, dependency graph, and transition matrix, are
shown.
,
its state space, dependency graph, and transition matrix, are
shown.
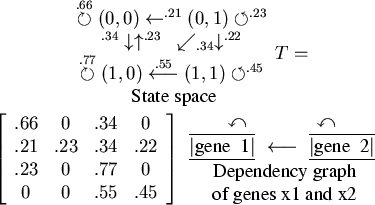 |
A mathematical method is constructed here given the dependency graph
of a set of genes. These genes could have either two, or three values.
The model that we obtain gives the
information about the subnetworks, the possible projections, and
the fixed points. We present here a methodology for construct discrete
networks using the dependency graph and a time series data.
For genes with more than two states, we assign three
possible values ![]() , and using the algorithm introduced in
[2] we calculate the model PRN.
, and using the algorithm introduced in
[2] we calculate the model PRN.
- 1
- M. A. Aviñó, "Homomorphisms of Probabilistic Gene Regulatory Networks", Proceedings of GENSIPS 2006.
- 2
- M. A. Aviñó, E. Green, and O. Moreno, "Applications of Finite Fields to Dynamical Systems and Reverse Engineering Problems", Proceedings of ACM Symposium on Applied Computing, 2004. doi:10.1145/967900.967939
- 3
- E. R. Dougherty, A. Datta, and C. Sima, "Developing therapeutic and diagnostic tools", Gen.Sig. Proc., IEEE Signal Processing Magazine, 46-68, Nov. 2005.
- 4
- I. Shmulevich, E. R. Dougherty, and W. Zhang, "From Boolean to probabilistic Boolean networks as models of genetic regulatory networks", Proc. of the IEEE. 90(11): 1778-1792, 2001. doi:10.1109/JPROC.2002.804686
purchase the full-text of this paper (price £20)
go to the previous paper
go to the next paper
return to the table of contents
return to the book description
purchase this book (price £105 +P&P)